 Nathanael Kuzio, NIH 2020-2022
Nathanael Kuzio, NIH 2020-2022
Graduate Program: Chemistry
Lab: Jeanne Hardy
Research Interests: Characterization and development of small molecule inhibitors of viral proteases through structure-activity relationship (SAR) optimization.
Research Summary
The sudden pandemics associated with novel viral outbreaks have contributed to over 50 million deaths since the beginning of the H1N1 pandemic in 1918. Historically, the unpredictability of these outbreaks has left us significantly unprepared, warranting robust research for future epidemics. In the latter half of the 20th century, substantial investigation into antiviral medicines has identified viral-encoded proteases, a component necessary for maturation of structural and nonstructural viral proteins, as a potential therapeutic target. Inhibition of the viral protease would stop proteolysis of the viral polyprotein and subsequent viral survival (Fig. 1). Therefore, viral protease inhibition has become a focus for the Hardy lab, whose research specializes in protease investigation and manipulation.
Recently, several viruses including zika and dengue of the flavivirus genus, have attracted attention due to zika virus’s sudden and detrimental outbreak in 2015. The Hardy lab is currently studying the nonstructural protein 3 (NS3) proteases, as well as their nonstructural protein 2B (NS2B) cofactors which together form the viral protease (NS2B-NS3 pro) for these two
viruses. Alongside this, we have been developing potential antiviral protease inhibitors using a drug development progression closely following that of many companies in the biotech
industry, including target validation, lead identification and optimization followed by further preclinical testing and drug candidate advancement. My research focuses on the development and characterization of novel enzyme inhibitors discovered in our lab for zika virus protease using both structure-activity relationship (SAR) guided design, incorporating X-ray protein crystallographic structures I have determined. Considering several of our nanomolar binding compounds, I have engaged in cocrystallization of lead compound analogue SM1 with zika virus protease, as well as computational modeling and docking studies. Furthermore, our lead compounds’ inability to inhibit the structurally and chemically similar dengue virus protease has fueled investigative efforts to identify divergent regions that could constitute binding to exclusively zika virus protease. This allows for insight into the requirements for both highly potent specific viral treatments, and pan-flaviviral therapeutics.
Throughout the course of my research, my medicinal chemistry studies are discussed with other members of the Hardy lab with varying areas of expertise, as well as other chemistry, chemical engineering, biochemistry, and molecular biology students and faculty through the UMass Joint Crystallography Group Meetings. From these collaborations, different viewpoints and opinions are brought forth weekly by individuals of varying backgrounds, feeding the planning of future experiments much like that of different teams within a biotech company. Ultimately, between the techniques and approaches learned from my colleagues and the skills that will be acquired from an industrial internship, I look forward to advancing my projects and making my contribution to the field of antiviral research.
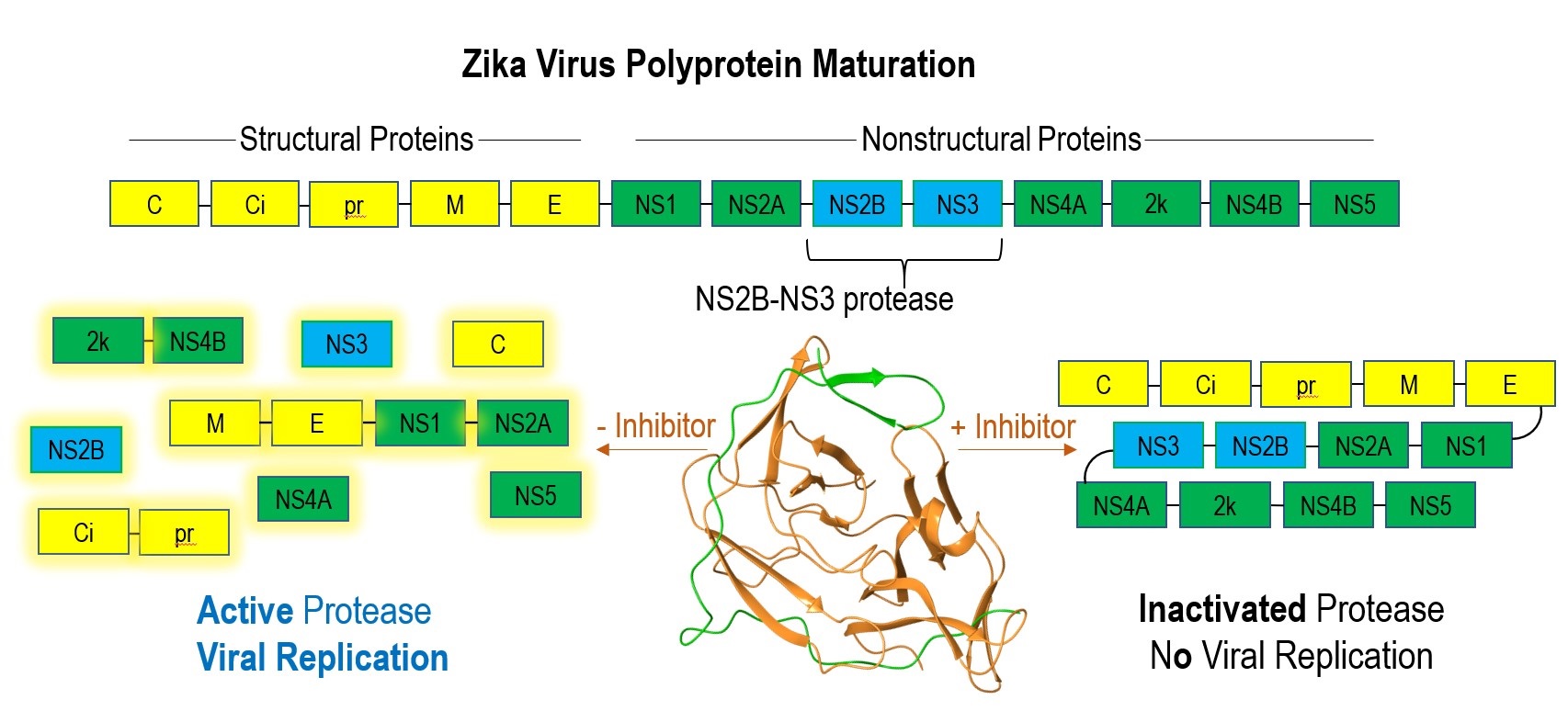 Figure 1. Maturation process of zika virus polyprotein with and without protease inhibitor. NS3 protease (orange), NS2B cofactor (green).
Figure 1. Maturation process of zika virus polyprotein with and without protease inhibitor. NS3 protease (orange), NS2B cofactor (green).
