Greg Gregory, NIH 2020-2022
Greg Gregory, NIH 2020-2022
Graduate Program: Plant Biology
Lab: Sam Hazen
Research Interests: Investigating gene regulation determinants to facilitate increased plant productivity through synthetic biology
Research Summary
All plant cells are surrounded by a wall that provides structure and protection. These walls are comprised mostly of polysaccharides, cellulose and hemicelluloses, as well as lignin, an extremely recalcitrant phenolic compound that cross-links with polysaccharides. Cell walls are a valuable commodity, providing textile fibers, pulp and paper, timber, as well as dietary fiber. Understanding the basic biological processes that create cell walls is a prerequisite for biotechnological strategies that could improve the quality, yield, and sustainability of plant derived products. Many of the enzymes that make cell wall polymers are characterized, as are several transcription factors that regulate wall growth and development. I propose to further dissect the protein-DNA interactions that result in cell wall biosynthesis for the purpose of developing a synthetic biology tool kit. With that we can design synthetic genes that can selectively drive endogenous pathways to increase plant growth and improve crop value.
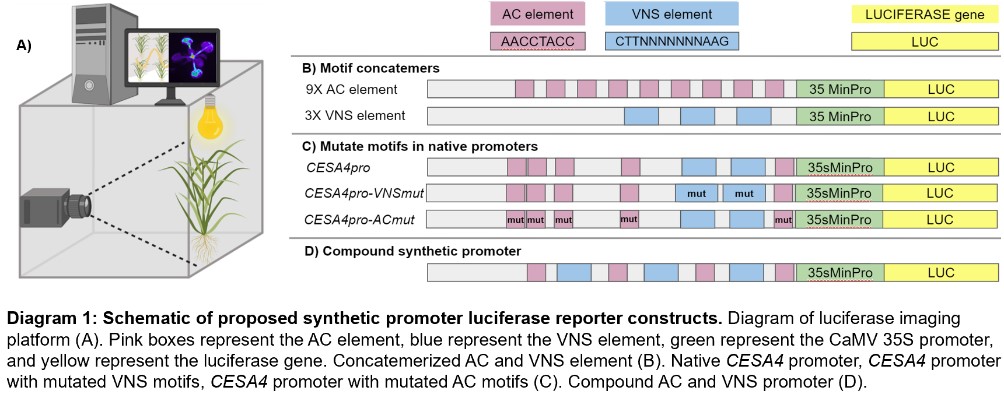
Cell wall thickening is in part controlled by transcription factor proteins that can bind specific DNA sequences, in particular the AC element (AACCTACC) and the VNS element (CTT*******AAG). The AC element interacts with several groups of MYB type proteins, while NAC type bind to the VNS element. While the function of these proteins has been described broadly, we understand very little about the dynamics of gene expression that result from these sequences and how they may interact to control wall synthesis. We want to better understand the functional role of the sequence motifs so that we can design synthetic genes that will have predictable expression behavior, enabling the development of designer agriculture applications.
In my project I will build synthetic gene promoters fused to a reporter gene, firefly luciferase. When expressed in our model organism Brachypodium distachyon and sprayed with Luciferin, activity of the reporter gene will result in light emission that I will measure with a CCD camera (Diagram 1A). The advantage of this system is that gene expression behavior, as determined by protein binding to the synthetic promoter, can be measured continuously and in any visible tissues under any conditions possible in the chamber. Images of bioluminescence will identify the levels, timing, and location of reporter gene expression. Three approaches will be used to test the functional role of the AC and VNS elements. First, I will build synthetic promoters that contain numerous copies of either the AC or VNS motif (Diagram 1B). Additionally, I will investigate the role of individual sequence motifs within native promoters, such as CELLULOSE SYNTHASE A4, by mutating the motifs within the promoter and observing the effect on reporter gene expression (Diagram 1C). Finally, I will construct compound synthetic promoters to investigate the role of motif combinations in gene expression (Diagram 1D). Once I characterize these sequence motifs to understand how they influence gene expression, they can be used in synthetic promoters to create specific spatiotemporal expression of any gene engineered in plants.