Akaansha Rampal, UMass Fellow 2021-2023
Akaansha Rampal, UMass Fellow 2021-2023
Graduate Program: Molecular and Cellular Biology
Lab: Shelly Peyton and Richard Vachet
Research Interests: Characterizing and comparing the distribution of extracellular matrix proteins in synthetic 3D hydrogels versus tissues
Research Summary
Introduction: Breast cancer is one of the leading causes of death in women. The extracellular matrix (ECM) is an important part of the tumor microenvironment and plays an important role in breast cancer invasion and metastasis. Intracellular biomarkers, such as proteins and miRNAs, have been used to predict tumor invasiveness, but there is a need to understand the role of ECM proteins in remodeling the matrix to promote metastatic growth. I use tunable polyethylene glycol-maleimide 3D hydrogels to mimic the complex tissue microenvironment by incorporating tissue-specific ECM components such as integrin binding peptides for cell adhesion and matrix metalloproteinases degradable peptide sequences for cell migration. These hydrogels polymerize faster than can be manually mixed, i.e. < 5 seconds, resulting in network heterogeneity and uneven particle distribution. However, we don’t yet know how polymerization rates affect the peptide distribution in these hydrogels. Our overarching goal is to compare the peptide distribution in our hydrogels versus healthy and diseased breast tissues. These distributions will give us insight into how accurately these hydrogels mimic the tissue microenvironment.
Our approach: To observe the peptide distributions in hydrogels and tissues I use two techniques:
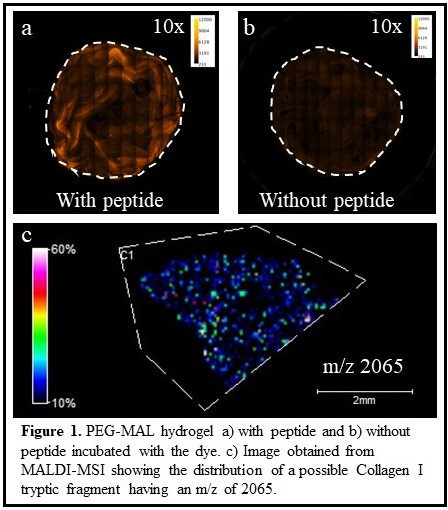
(1) To visualize peptide arrangement in hydrogels, I utilize fluorescent microscopy. I tag the integrin binding peptide (GPR) with a fluorescent dye, rhodamine B isothiocyanate. The dye reacts with the primary amine on the peptide forming a covalent amide bond. To avoid any changes in the gelation kinetics, I add the fluorescent dye after the hydrogels were formed. Our preliminary data show a homogenous distribution of the GPR peptide in our hydrogels (Figure 1a, b). However, these hydrogels contain more than one peptide and until now, I have successfully optimized the conditions to visualize a single peptide. I consider tagging the other peptides in the gels with different dyes to image them and further investigate how the peptide distribution in the hydrogel changes with varying gelation speeds.
(2) To visualize ECM protein distributions in tissues, I explore matrix-assisted laser desorption ionization mass spectrometry imaging (MALDI-MSI). MALDI-MSI generates molecular snapshots of the sample in a rapid and multiplexed manner and provides a map of the distributions of the biomolecules detected including lipids, peptides, and proteins without any chemical modification of the sample. I am measuring the distributions of ECM proteins in tissues by using tryptic peptide fragments as indicators of these proteins. Figure 1c shows the distribution of a tryptic peptide fragment in liver tissue, having a m/z of 2065, which is likely derived from the Collagen I protein. I have detected many additional tryptic fragments, but MS/MS analysis is needed to confirm the identities of the peptides (and their corresponding) proteins. Using this approach, I can image the spatial distribution of many ECM proteins in a single experiment